Alternating Poisson Regression for fitting CP to sparse count data
References:
- E. C. Chi, T. G. Kolda, On Tensors, Sparsity, and Nonnegative Factorizations, SIAM J. Matrix Analysis and Applications, 33:1272-1299, 2012, https://doi.org/10.1137/110859063
- S. Hansen, T. Plantenga and T. G. Kolda, Newton-Based Optimization for Kullback-Leibler Nonnegative Tensor Factorizations, Optimization Methods and Software, 30(5):955-979, 2015, http://dx.doi.org/10.1080/10556788.2015.1009977
Contents
Set up a sample problem
We follow the general procedure for creating a problem outlined by Chi and Kolda (2012). This creates a sparse count tensor with a known solution. The solution is a CP decomposition with a few large entries in each column of the factor matrices. The solution is normalized and sorted by component size in descending order.
rng('default') %<- Setting random seed for reproducibility of this script % Pick the size and rank sz = [100 80 60]; R = 5; % Generate factor matrices with a few large entries in each column; this % will be the basis of our soln. A = cell(3,1); for n = 1:length(sz) A{n} = rand(sz(n), R); for r = 1:R p = randperm(sz(n)); nbig = round( (1/R)*sz(n) ); A{n}(p(1:nbig),r) = 100 * A{n}(p(1:nbig),r); end end lambda = rand(R,1); S = ktensor(lambda, A); S = normalize(S,'sort',1); % Create sparse test problem based on provided solution. nz = prod(sz) * .05; info = create_problem('Soln', S, 'Sparse_Generation', nz); % Extract data and solution X = info.Data; M_true = info.Soln;
Call CP-APR
Alternating Poisson Regression (APR) is a method for fitting a CP decomposition to sparse count data. It is a nonnegative method that minimizes the Kullback-Leibler divergence between the data and the model. The method is implemented in the cp_apr function, which is similar to the cp_als function, but uses a different objective function and optimization method. The cp_apr function is designed to handle sparse count data and is particularly useful for fitting nonnegative CP decompositions.
The cp_apr function is a wrapper that calls one of three specific algorithms, selected by the 'alg' parameter:
- 'pqnr' (Default): Row subproblems are solved by Projected Quasi-Newton with L-BFGS. This method generally offers a good balance of speed and robustness and is suitable for a wide range of problems. It approximates the Hessian using gradient information from previous iterations. It is based on the work by Hansen, Plantenga, and Kolda (2015).
- 'pdnr': Row subproblems are solved by Projected Damped Newton. This method uses the exact Hessian for the row subproblems, which can lead to higher accuracy per iteration but may be more computationally intensive, especially for large R, as it involves forming and solving an R x R system at each inner iteration for each row. It is based on the work by Hansen, Plantenga, and Kolda (2015).
- 'mu': Multiplicative Update. This is a simpler algorithm, often with cheaper iterations. It can be slower to converge to high accuracy compared to Newton-based methods but can be effective for very large, sparse problems or for obtaining an initial guess quickly. It is based on the work by Chi & Kolda (2012).
The following example uses the default 'pqnr' algorithm. We run the method 5 times and keep the best solution (lowest objective value).
% Compute a solution using the default 'pqnr' algorithm fprintf('--- Running CP-APR with PQNR (default) ---\n'); rng('default') best_obj = inf; for trial = 1:5 [M_tmp,~,output_tmp] = cp_apr(X, R, 'printitn', 2); if output_tmp.obj < best_obj best_obj = output_tmp.obj; M_pqnr = M_tmp; output_pqnr = output_tmp; end end % Score the solution (a score of 1 is perfect) factor_match_score_pqnr = score(M_pqnr, M_true, 'greedy', true);
--- Running CP-APR with PQNR (default) --- CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pqnr' (quasi-Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 2370, KKT viol = 3.72e+00, obj = 1.64937774e+04, nz: 271 4. Ttl Inner Its: 2164, KKT viol = 1.81e+00, obj = 1.26971895e+04, nz: 249 6. Ttl Inner Its: 1742, KKT viol = 5.75e-01, obj = 1.25597728e+04, nz: 276 8. Ttl Inner Its: 1148, KKT viol = 4.01e-02, obj = 1.25592937e+04, nz: 279 10. Ttl Inner Its: 488, KKT viol = 7.84e-03, obj = 1.25592875e+04, nz: 280 12. Ttl Inner Its: 293, KKT viol = 3.86e-03, obj = 1.25592870e+04, nz: 280 14. Ttl Inner Its: 273, KKT viol = 1.96e-03, obj = 1.25592869e+04, nz: 280 16. Ttl Inner Its: 249, KKT viol = 9.17e-04, obj = 1.25592869e+04, nz: 280 18. Ttl Inner Its: 246, KKT viol = 4.78e-04, obj = 1.25592869e+04, nz: 280 20. Ttl Inner Its: 242, KKT viol = 1.94e-04, obj = 1.25592869e+04, nz: 280 22. Ttl Inner Its: 241, KKT viol = 1.09e-04, obj = 1.25592869e+04, nz: 280 =========================================== Final f = 1.255929e+04 Final least squares fit = 5.232288e-01 Final KKT violation = 9.8366707e-05 Total inner iterations = 20219 Total execution time = 1.12 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pqnr' (quasi-Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 2364, KKT viol = 4.77e+00, obj = 1.74548483e+04, nz: 321 4. Ttl Inner Its: 2282, KKT viol = 2.35e+00, obj = 1.48353911e+04, nz: 283 6. Ttl Inner Its: 2083, KKT viol = 4.68e-01, obj = 1.44062370e+04, nz: 276 8. Ttl Inner Its: 1814, KKT viol = 1.11e-01, obj = 1.43840303e+04, nz: 287 10. Ttl Inner Its: 1649, KKT viol = 2.49e-01, obj = 1.43731087e+04, nz: 290 12. Ttl Inner Its: 1579, KKT viol = 1.75e-01, obj = 1.43671444e+04, nz: 291 14. Ttl Inner Its: 1304, KKT viol = 6.69e-02, obj = 1.43656031e+04, nz: 295 16. Ttl Inner Its: 1338, KKT viol = 1.50e-01, obj = 1.43641731e+04, nz: 293 18. Ttl Inner Its: 1355, KKT viol = 6.13e-02, obj = 1.43621016e+04, nz: 296 20. Ttl Inner Its: 1264, KKT viol = 4.25e-02, obj = 1.43611481e+04, nz: 302 22. Ttl Inner Its: 953, KKT viol = 4.40e-02, obj = 1.43609727e+04, nz: 306 24. Ttl Inner Its: 894, KKT viol = 3.46e-02, obj = 1.43608920e+04, nz: 306 26. Ttl Inner Its: 1007, KKT viol = 3.55e-02, obj = 1.43608107e+04, nz: 306 28. Ttl Inner Its: 865, KKT viol = 5.94e-03, obj = 1.43607903e+04, nz: 307 30. Ttl Inner Its: 724, KKT viol = 3.37e-03, obj = 1.43607868e+04, nz: 306 32. Ttl Inner Its: 552, KKT viol = 1.94e-03, obj = 1.43607858e+04, nz: 306 34. Ttl Inner Its: 414, KKT viol = 8.77e-04, obj = 1.43607854e+04, nz: 306 36. Ttl Inner Its: 314, KKT viol = 5.43e-04, obj = 1.43607853e+04, nz: 306 38. Ttl Inner Its: 258, KKT viol = 1.82e-04, obj = 1.43607853e+04, nz: 306 40. Ttl Inner Its: 247, KKT viol = 1.23e-04, obj = 1.43607853e+04, nz: 306 =========================================== Final f = 1.436079e+04 Final least squares fit = 5.160188e-01 Final KKT violation = 9.5163112e-05 Total inner iterations = 47847 Total execution time = 2.11 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pqnr' (quasi-Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 2354, KKT viol = 6.59e+00, obj = 1.37026077e+04, nz: 263 4. Ttl Inner Its: 1997, KKT viol = 7.56e-01, obj = 1.25681246e+04, nz: 267 6. Ttl Inner Its: 1567, KKT viol = 1.62e-01, obj = 1.25599271e+04, nz: 278 8. Ttl Inner Its: 698, KKT viol = 3.90e-02, obj = 1.25598567e+04, nz: 278 10. Ttl Inner Its: 464, KKT viol = 2.07e-02, obj = 1.25598465e+04, nz: 278 12. Ttl Inner Its: 313, KKT viol = 1.07e-02, obj = 1.25598437e+04, nz: 279 14. Ttl Inner Its: 314, KKT viol = 5.39e-03, obj = 1.25598430e+04, nz: 279 16. Ttl Inner Its: 263, KKT viol = 2.70e-03, obj = 1.25598428e+04, nz: 279 18. Ttl Inner Its: 254, KKT viol = 1.36e-03, obj = 1.25598428e+04, nz: 279 20. Ttl Inner Its: 248, KKT viol = 6.69e-04, obj = 1.25598428e+04, nz: 279 22. Ttl Inner Its: 247, KKT viol = 3.47e-04, obj = 1.25598428e+04, nz: 279 24. Ttl Inner Its: 242, KKT viol = 1.40e-04, obj = 1.25598428e+04, nz: 279 26. Ttl Inner Its: 240, KKT viol = 9.82e-05, obj = 1.25598427e+04, nz: 279 =========================================== Final f = 1.255984e+04 Final least squares fit = 5.232265e-01 Final KKT violation = 9.8211173e-05 Total inner iterations = 19446 Total execution time = 1.14 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pqnr' (quasi-Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 2359, KKT viol = 3.34e+04, obj = 1.54901159e+04, nz: 266 4. Ttl Inner Its: 2119, KKT viol = 1.00e+00, obj = 1.44274757e+04, nz: 283 6. Ttl Inner Its: 1812, KKT viol = 2.61e-01, obj = 1.43754371e+04, nz: 288 8. Ttl Inner Its: 1586, KKT viol = 1.98e-01, obj = 1.43684214e+04, nz: 293 10. Ttl Inner Its: 1478, KKT viol = 1.37e-01, obj = 1.43646587e+04, nz: 299 12. Ttl Inner Its: 1411, KKT viol = 1.96e-01, obj = 1.43596863e+04, nz: 303 14. Ttl Inner Its: 1467, KKT viol = 2.05e-01, obj = 1.43566765e+04, nz: 296 16. Ttl Inner Its: 1660, KKT viol = 3.69e-01, obj = 1.43401947e+04, nz: 286 18. Ttl Inner Its: 2019, KKT viol = 2.19e+00, obj = 1.33546642e+04, nz: 265 20. Ttl Inner Its: 1891, KKT viol = 9.14e-01, obj = 1.25614742e+04, nz: 274 22. Ttl Inner Its: 1062, KKT viol = 3.59e-02, obj = 1.25593464e+04, nz: 279 24. Ttl Inner Its: 439, KKT viol = 8.91e-03, obj = 1.25593396e+04, nz: 280 26. Ttl Inner Its: 312, KKT viol = 4.40e-03, obj = 1.25593389e+04, nz: 281 28. Ttl Inner Its: 277, KKT viol = 2.27e-03, obj = 1.25593388e+04, nz: 281 30. Ttl Inner Its: 250, KKT viol = 1.09e-03, obj = 1.25593388e+04, nz: 281 32. Ttl Inner Its: 244, KKT viol = 5.31e-04, obj = 1.25593388e+04, nz: 281 34. Ttl Inner Its: 244, KKT viol = 2.94e-04, obj = 1.25593388e+04, nz: 281 36. Ttl Inner Its: 243, KKT viol = 1.25e-04, obj = 1.25593388e+04, nz: 281 =========================================== Final f = 1.255934e+04 Final least squares fit = 5.232254e-01 Final KKT violation = 9.5937950e-05 Total inner iterations = 42943 Total execution time = 2.20 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pqnr' (quasi-Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 2362, KKT viol = 4.33e+00, obj = 1.61058863e+04, nz: 261 4. Ttl Inner Its: 2187, KKT viol = 1.78e+00, obj = 1.30304826e+04, nz: 264 6. Ttl Inner Its: 1922, KKT viol = 9.02e-01, obj = 1.25620840e+04, nz: 273 8. Ttl Inner Its: 1422, KKT viol = 8.22e-02, obj = 1.25593143e+04, nz: 278 10. Ttl Inner Its: 524, KKT viol = 1.25e-02, obj = 1.25592873e+04, nz: 280 12. Ttl Inner Its: 303, KKT viol = 2.70e-03, obj = 1.25592869e+04, nz: 280 14. Ttl Inner Its: 258, KKT viol = 1.36e-03, obj = 1.25592869e+04, nz: 280 16. Ttl Inner Its: 252, KKT viol = 6.63e-04, obj = 1.25592869e+04, nz: 280 18. Ttl Inner Its: 244, KKT viol = 3.32e-04, obj = 1.25592869e+04, nz: 280 20. Ttl Inner Its: 244, KKT viol = 1.52e-04, obj = 1.25592869e+04, nz: 280 =========================================== Final f = 1.255929e+04 Final least squares fit = 5.232290e-01 Final KKT violation = 9.5799877e-05 Total inner iterations = 20721 Total execution time = 1.13 secs
Example using the 'pdnr' algorithm
Here, we explicitly select the 'pdnr' algorithm. We also reduce the maximum number of iterations for this demonstration. We run the method 5 times and keep the best solution (lowest objective value).
fprintf('--- Running CP-APR with PDNR ---\n'); rng('default') best_obj_pdnr = inf; for trial = 1:5 [M_tmp,~,output_tmp] = cp_apr(X, R, 'alg', 'pdnr', 'printitn', 2); if output_tmp.obj < best_obj_pdnr best_obj_pdnr = output_tmp.obj; M_pdnr = M_tmp; output_pdnr = output_tmp; end end % Score the solution factor_match_score_pdnr = score(M_pdnr, M_true, 'greedy', true);
--- Running CP-APR with PDNR --- CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pdnr' (damped Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 mu0 = 1e-05, inexact = 1 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 1224, KKT viol = 8.10e+02, obj = 2.08873010e+04, nz: 340 4. Ttl Inner Its: 240, KKT viol = 7.86e+00, obj = 2.04610219e+04, nz: 309 6. Ttl Inner Its: 294, KKT viol = 4.67e+01, obj = 1.73189096e+04, nz: 307 8. Ttl Inner Its: 640, KKT viol = 3.51e+00, obj = 1.66981705e+04, nz: 308 10. Ttl Inner Its: 263, KKT viol = 5.21e+01, obj = 1.45105514e+04, nz: 279 12. Ttl Inner Its: 544, KKT viol = 1.89e+00, obj = 1.43716675e+04, nz: 297 14. Ttl Inner Its: 391, KKT viol = 2.31e-01, obj = 1.43653907e+04, nz: 297 16. Ttl Inner Its: 431, KKT viol = 4.82e-02, obj = 1.43640168e+04, nz: 298 18. Ttl Inner Its: 478, KKT viol = 2.09e-01, obj = 1.43626771e+04, nz: 300 20. Ttl Inner Its: 470, KKT viol = 2.70e-02, obj = 1.43617928e+04, nz: 301 22. Ttl Inner Its: 464, KKT viol = 6.34e-02, obj = 1.43611589e+04, nz: 301 24. Ttl Inner Its: 465, KKT viol = 3.30e-02, obj = 1.43605962e+04, nz: 304 26. Ttl Inner Its: 474, KKT viol = 2.68e-02, obj = 1.43603417e+04, nz: 301 28. Ttl Inner Its: 473, KKT viol = 8.16e-02, obj = 1.43601599e+04, nz: 299 30. Ttl Inner Its: 440, KKT viol = 1.03e-01, obj = 1.43596476e+04, nz: 301 32. Ttl Inner Its: 451, KKT viol = 8.07e-02, obj = 1.43590520e+04, nz: 302 34. Ttl Inner Its: 464, KKT viol = 2.28e-01, obj = 1.43580597e+04, nz: 302 36. Ttl Inner Its: 484, KKT viol = 4.16e-02, obj = 1.43576430e+04, nz: 300 38. Ttl Inner Its: 474, KKT viol = 4.23e-02, obj = 1.43572954e+04, nz: 299 40. Ttl Inner Its: 478, KKT viol = 4.78e-02, obj = 1.43569118e+04, nz: 295 42. Ttl Inner Its: 489, KKT viol = 1.14e-01, obj = 1.43557419e+04, nz: 295 44. Ttl Inner Its: 505, KKT viol = 1.51e-01, obj = 1.43524794e+04, nz: 292 46. Ttl Inner Its: 471, KKT viol = 1.04e+00, obj = 1.43477325e+04, nz: 292 48. Ttl Inner Its: 408, KKT viol = 2.67e-02, obj = 1.43466268e+04, nz: 294 50. Ttl Inner Its: 470, KKT viol = 2.72e-02, obj = 1.43463649e+04, nz: 294 52. Ttl Inner Its: 468, KKT viol = 2.60e-02, obj = 1.43462201e+04, nz: 294 54. Ttl Inner Its: 465, KKT viol = 2.40e-02, obj = 1.43461180e+04, nz: 293 56. Ttl Inner Its: 471, KKT viol = 1.54e-02, obj = 1.43460584e+04, nz: 293 58. Ttl Inner Its: 469, KKT viol = 1.49e-02, obj = 1.43460250e+04, nz: 292 60. Ttl Inner Its: 463, KKT viol = 1.45e-02, obj = 1.43459990e+04, nz: 292 62. Ttl Inner Its: 469, KKT viol = 9.39e-03, obj = 1.43459796e+04, nz: 293 64. Ttl Inner Its: 466, KKT viol = 6.91e-03, obj = 1.43459683e+04, nz: 293 66. Ttl Inner Its: 462, KKT viol = 5.11e-03, obj = 1.43459622e+04, nz: 293 68. Ttl Inner Its: 462, KKT viol = 3.63e-03, obj = 1.43459589e+04, nz: 292 70. Ttl Inner Its: 459, KKT viol = 2.55e-03, obj = 1.43459570e+04, nz: 292 72. Ttl Inner Its: 454, KKT viol = 1.80e-03, obj = 1.43459559e+04, nz: 292 74. Ttl Inner Its: 437, KKT viol = 1.20e-03, obj = 1.43459553e+04, nz: 292 76. Ttl Inner Its: 392, KKT viol = 8.44e-04, obj = 1.43459550e+04, nz: 292 78. Ttl Inner Its: 345, KKT viol = 4.35e-04, obj = 1.43459549e+04, nz: 292 80. Ttl Inner Its: 282, KKT viol = 2.61e-04, obj = 1.43459548e+04, nz: 292 82. Ttl Inner Its: 264, KKT viol = 2.27e-04, obj = 1.43459548e+04, nz: 292 84. Ttl Inner Its: 271, KKT viol = 3.03e-04, obj = 1.43459547e+04, nz: 292 86. Ttl Inner Its: 240, KKT viol = 9.94e-05, obj = 1.43459547e+04, nz: 292 =========================================== Final f = 1.434595e+04 Final least squares fit = 5.147296e-01 Final KKT violation = 9.9407836e-05 Total inner iterations = 39250 Total execution time = 4.40 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pdnr' (damped Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 mu0 = 1e-05, inexact = 1 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 620, KKT viol = 1.87e+03, obj = 3.72481432e+04, nz: 327 4. Ttl Inner Its: 1082, KKT viol = 5.42e+01, obj = 1.71493242e+04, nz: 325 6. Ttl Inner Its: 245, KKT viol = 6.15e+01, obj = 1.30919319e+04, nz: 263 8. Ttl Inner Its: 643, KKT viol = 9.41e-01, obj = 1.25717033e+04, nz: 275 10. Ttl Inner Its: 300, KKT viol = 4.10e-01, obj = 1.25598716e+04, nz: 278 12. Ttl Inner Its: 325, KKT viol = 4.34e-02, obj = 1.25597842e+04, nz: 279 14. Ttl Inner Its: 335, KKT viol = 1.30e-02, obj = 1.25597785e+04, nz: 279 16. Ttl Inner Its: 303, KKT viol = 4.89e-03, obj = 1.25597781e+04, nz: 280 18. Ttl Inner Its: 253, KKT viol = 2.41e-03, obj = 1.25597781e+04, nz: 280 20. Ttl Inner Its: 251, KKT viol = 1.20e-03, obj = 1.25597781e+04, nz: 280 22. Ttl Inner Its: 244, KKT viol = 5.73e-04, obj = 1.25597781e+04, nz: 280 24. Ttl Inner Its: 244, KKT viol = 2.64e-04, obj = 1.25597781e+04, nz: 280 26. Ttl Inner Its: 244, KKT viol = 1.50e-04, obj = 1.25597781e+04, nz: 280 28. Ttl Inner Its: 240, KKT viol = 9.97e-05, obj = 1.25597781e+04, nz: 280 =========================================== Final f = 1.255978e+04 Final least squares fit = 5.232344e-01 Final KKT violation = 9.9668454e-05 Total inner iterations = 10401 Total execution time = 1.08 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pdnr' (damped Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 mu0 = 1e-05, inexact = 1 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 521, KKT viol = 1.58e+03, obj = 3.57843712e+04, nz: 325 4. Ttl Inner Its: 1013, KKT viol = 4.47e+01, obj = 1.83420068e+04, nz: 342 6. Ttl Inner Its: 240, KKT viol = 2.83e+00, obj = 1.71814960e+04, nz: 296 8. Ttl Inner Its: 524, KKT viol = 1.71e+00, obj = 1.68610010e+04, nz: 318 10. Ttl Inner Its: 727, KKT viol = 2.25e+02, obj = 1.44014052e+04, nz: 290 12. Ttl Inner Its: 463, KKT viol = 2.06e+00, obj = 1.42891949e+04, nz: 287 14. Ttl Inner Its: 802, KKT viol = 4.34e+01, obj = 1.26000219e+04, nz: 269 16. Ttl Inner Its: 371, KKT viol = 6.95e-01, obj = 1.25640961e+04, nz: 278 18. Ttl Inner Its: 328, KKT viol = 1.76e-01, obj = 1.25593059e+04, nz: 278 20. Ttl Inner Its: 346, KKT viol = 1.42e-02, obj = 1.25592875e+04, nz: 280 22. Ttl Inner Its: 322, KKT viol = 6.92e-03, obj = 1.25592870e+04, nz: 280 24. Ttl Inner Its: 261, KKT viol = 3.43e-03, obj = 1.25592869e+04, nz: 280 26. Ttl Inner Its: 247, KKT viol = 1.68e-03, obj = 1.25592868e+04, nz: 280 28. Ttl Inner Its: 246, KKT viol = 7.81e-04, obj = 1.25592868e+04, nz: 280 30. Ttl Inner Its: 245, KKT viol = 3.78e-04, obj = 1.25592868e+04, nz: 280 32. Ttl Inner Its: 243, KKT viol = 1.32e-04, obj = 1.25592868e+04, nz: 280 34. Ttl Inner Its: 240, KKT viol = 9.98e-05, obj = 1.25592868e+04, nz: 280 =========================================== Final f = 1.255929e+04 Final least squares fit = 5.232291e-01 Final KKT violation = 9.9761257e-05 Total inner iterations = 14152 Total execution time = 1.36 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pdnr' (damped Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 mu0 = 1e-05, inexact = 1 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 324, KKT viol = 1.09e+03, obj = 5.90744136e+04, nz: 318 4. Ttl Inner Its: 1205, KKT viol = 1.48e+03, obj = 2.03017275e+04, nz: 372 6. Ttl Inner Its: 240, KKT viol = 3.24e+01, obj = 1.94325932e+04, nz: 326 8. Ttl Inner Its: 487, KKT viol = 1.07e+02, obj = 1.33200711e+04, nz: 249 10. Ttl Inner Its: 872, KKT viol = 1.90e+01, obj = 1.26266158e+04, nz: 256 12. Ttl Inner Its: 264, KKT viol = 9.34e-01, obj = 1.25675319e+04, nz: 270 14. Ttl Inner Its: 432, KKT viol = 3.45e-01, obj = 1.25598890e+04, nz: 276 16. Ttl Inner Its: 273, KKT viol = 9.89e-02, obj = 1.25597954e+04, nz: 277 18. Ttl Inner Its: 433, KKT viol = 6.45e-03, obj = 1.25597900e+04, nz: 278 20. Ttl Inner Its: 278, KKT viol = 3.51e-03, obj = 1.25597899e+04, nz: 278 22. Ttl Inner Its: 248, KKT viol = 1.75e-03, obj = 1.25597899e+04, nz: 278 24. Ttl Inner Its: 244, KKT viol = 8.24e-04, obj = 1.25597899e+04, nz: 278 26. Ttl Inner Its: 246, KKT viol = 3.94e-04, obj = 1.25597899e+04, nz: 278 28. Ttl Inner Its: 247, KKT viol = 2.11e-04, obj = 1.25597899e+04, nz: 278 30. Ttl Inner Its: 242, KKT viol = 1.12e-04, obj = 1.25597899e+04, nz: 278 =========================================== Final f = 1.255979e+04 Final least squares fit = 5.232301e-01 Final KKT violation = 9.9705252e-05 Total inner iterations = 11786 Total execution time = 1.38 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'pdnr' (damped Newton) stoptol = 0.0001, stoptime = 1e+06 maxiters = 1000, maxinneriters = 10 epsDivZero = 1e-10, epsActive = 1e-08 mu0 = 1e-05, inexact = 1 precompinds = 1 Precomputing sparse index sets...done 2. Ttl Inner Its: 578, KKT viol = 7.88e+02, obj = 3.67776143e+04, nz: 282 4. Ttl Inner Its: 334, KKT viol = 1.48e+01, obj = 3.34715128e+04, nz: 256 6. Ttl Inner Its: 647, KKT viol = 1.55e+02, obj = 1.53709982e+04, nz: 267 8. Ttl Inner Its: 907, KKT viol = 2.37e+00, obj = 1.47265281e+04, nz: 271 10. Ttl Inner Its: 490, KKT viol = 2.50e+01, obj = 1.27458113e+04, nz: 257 12. Ttl Inner Its: 429, KKT viol = 2.23e+00, obj = 1.25642707e+04, nz: 271 14. Ttl Inner Its: 385, KKT viol = 1.29e-01, obj = 1.25593488e+04, nz: 277 16. Ttl Inner Its: 356, KKT viol = 3.29e-02, obj = 1.25592907e+04, nz: 279 18. Ttl Inner Its: 319, KKT viol = 1.73e-02, obj = 1.25592877e+04, nz: 280 20. Ttl Inner Its: 303, KKT viol = 8.70e-03, obj = 1.25592871e+04, nz: 280 22. Ttl Inner Its: 260, KKT viol = 4.46e-03, obj = 1.25592869e+04, nz: 280 24. Ttl Inner Its: 252, KKT viol = 2.23e-03, obj = 1.25592868e+04, nz: 280 26. Ttl Inner Its: 252, KKT viol = 1.14e-03, obj = 1.25592868e+04, nz: 280 28. Ttl Inner Its: 249, KKT viol = 5.76e-04, obj = 1.25592868e+04, nz: 280 30. Ttl Inner Its: 244, KKT viol = 2.90e-04, obj = 1.25592868e+04, nz: 280 32. Ttl Inner Its: 243, KKT viol = 1.58e-04, obj = 1.25592868e+04, nz: 280 34. Ttl Inner Its: 240, KKT viol = 9.97e-05, obj = 1.25592868e+04, nz: 280 =========================================== Final f = 1.255929e+04 Final least squares fit = 5.232289e-01 Final KKT violation = 9.9691480e-05 Total inner iterations = 13453 Total execution time = 1.28 secs
Example using the 'mu' algorithm
This example demonstrates the 'mu' algorithm. We can also set parameters specific to 'mu', like 'kappa'. Because this is faster per iteration by requires more iterations, we only print the results every 10 iterations. We run the method 5 times and keep the best solution (lowest objective value).
fprintf('--- Running CP-APR with MU ---\n'); rng('default') best_obj_mu = inf; for trial = 1:5 [M_tmp,~,output_tmp] = cp_apr(X, R, 'alg', 'mu', 'printitn', 10, 'maxiters', 200, 'kappa', 50); if output_tmp.obj < best_obj_mu best_obj_mu = output_tmp.obj; M_mu = M_tmp; output_mu = output_tmp; end end % Score the solution factor_match_score_mu = score(M_mu, M_true, 'greedy', true);
--- Running CP-APR with MU --- CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'mu' (multiplicative updates with adjustments) stoptol = 0.0001, stoptime = 1e+06 maxiters = 200, maxinneriters = 10 epsDivZero = 1e-10, kappa = 50, kappatol = 1e-10 Iter 10: Inner Its = 30 KKT violation = 9.249377e-02, nViolations = 0, obj = 1.25598982e+04 Iter 20: Inner Its = 30 KKT violation = 8.592690e-03, nViolations = 0, obj = 1.25597790e+04 Iter 30: Inner Its = 24 KKT violation = 5.806821e-03, nViolations = 0, obj = 1.25597782e+04 Iter 40: Inner Its = 15 KKT violation = 4.655408e-03, nViolations = 0, obj = 1.25597781e+04 Iter 50: Inner Its = 12 KKT violation = 4.025659e-03, nViolations = 0, obj = 1.25597781e+04 Iter 60: Inner Its = 12 KKT violation = 3.497964e-03, nViolations = 0, obj = 1.25597781e+04 Iter 70: Inner Its = 12 KKT violation = 2.457578e-03, nViolations = 0, obj = 1.25597781e+04 Iter 80: Inner Its = 12 KKT violation = 1.759166e-03, nViolations = 0, obj = 1.25597781e+04 Iter 90: Inner Its = 12 KKT violation = 1.277045e-03, nViolations = 0, obj = 1.25597781e+04 Iter 100: Inner Its = 12 KKT violation = 9.350392e-04, nViolations = 0, obj = 1.25597781e+04 Iter 110: Inner Its = 12 KKT violation = 6.898427e-04, nViolations = 0, obj = 1.25597781e+04 Iter 120: Inner Its = 12 KKT violation = 5.106160e-04, nViolations = 0, obj = 1.25597781e+04 Iter 130: Inner Its = 12 KKT violation = 3.798073e-04, nViolations = 0, obj = 1.25597781e+04 Iter 140: Inner Its = 12 KKT violation = 2.824650e-04, nViolations = 0, obj = 1.25597781e+04 Iter 150: Inner Its = 12 KKT violation = 2.108602e-04, nViolations = 0, obj = 1.25597781e+04 Iter 160: Inner Its = 12 KKT violation = 1.578435e-04, nViolations = 0, obj = 1.25597781e+04 Iter 170: Inner Its = 12 KKT violation = 1.183998e-04, nViolations = 0, obj = 1.25597781e+04 Exiting because all subproblems reached KKT tol. =========================================== Final f = 1.255978e+04 Final least squares fit = 5.232344e-01 Final KKT violation = 9.9999544e-05 Total inner iterations = 2723 Total execution time = 1.62 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'mu' (multiplicative updates with adjustments) stoptol = 0.0001, stoptime = 1e+06 maxiters = 200, maxinneriters = 10 epsDivZero = 1e-10, kappa = 50, kappatol = 1e-10 Iter 10: Inner Its = 30 KKT violation = 4.844923e-01, nViolations = 2, obj = 1.43860744e+04 Iter 20: Inner Its = 30 KKT violation = 1.738390e-01, nViolations = 1, obj = 1.43648752e+04 Iter 30: Inner Its = 30 KKT violation = 7.799808e-02, nViolations = 0, obj = 1.43609810e+04 Iter 40: Inner Its = 30 KKT violation = 1.032798e-01, nViolations = 0, obj = 1.43595585e+04 Iter 50: Inner Its = 30 KKT violation = 4.356253e-02, nViolations = 0, obj = 1.43592219e+04 Iter 60: Inner Its = 30 KKT violation = 2.071341e-02, nViolations = 0, obj = 1.43591517e+04 Iter 70: Inner Its = 30 KKT violation = 7.858970e-03, nViolations = 0, obj = 1.43591337e+04 Iter 80: Inner Its = 28 KKT violation = 7.752428e-03, nViolations = 0, obj = 1.43591298e+04 Iter 90: Inner Its = 30 KKT violation = 7.783842e-03, nViolations = 0, obj = 1.43591274e+04 Iter 100: Inner Its = 30 KKT violation = 7.986604e-03, nViolations = 0, obj = 1.43591236e+04 Iter 110: Inner Its = 30 KKT violation = 8.409126e-03, nViolations = 0, obj = 1.43591173e+04 Iter 120: Inner Its = 30 KKT violation = 8.402947e-03, nViolations = 0, obj = 1.43591128e+04 Iter 130: Inner Its = 30 KKT violation = 3.890598e-03, nViolations = 0, obj = 1.43591073e+04 Iter 140: Inner Its = 30 KKT violation = 3.769377e-03, nViolations = 0, obj = 1.43590991e+04 Iter 150: Inner Its = 30 KKT violation = 3.365400e-02, nViolations = 0, obj = 1.43590918e+04 Iter 160: Inner Its = 30 KKT violation = 4.143578e-03, nViolations = 0, obj = 1.43590671e+04 Iter 170: Inner Its = 30 KKT violation = 3.009754e-03, nViolations = 0, obj = 1.43590386e+04 Iter 180: Inner Its = 30 KKT violation = 4.864634e-03, nViolations = 0, obj = 1.43589935e+04 Iter 190: Inner Its = 30 KKT violation = 1.358162e-01, nViolations = 1, obj = 1.43611969e+04 Iter 200: Inner Its = 30 KKT violation = 3.583451e-02, nViolations = 0, obj = 1.43575841e+04 =========================================== Final f = 1.435758e+04 Final least squares fit = 5.164555e-01 Final KKT violation = 3.5834509e-02 Total inner iterations = 5992 Total execution time = 3.03 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'mu' (multiplicative updates with adjustments) stoptol = 0.0001, stoptime = 1e+06 maxiters = 200, maxinneriters = 10 epsDivZero = 1e-10, kappa = 50, kappatol = 1e-10 Iter 10: Inner Its = 30 KKT violation = 4.074251e-02, nViolations = 0, obj = 1.25598024e+04 Iter 20: Inner Its = 30 KKT violation = 7.985892e-03, nViolations = 0, obj = 1.25597901e+04 Iter 30: Inner Its = 21 KKT violation = 5.531637e-03, nViolations = 0, obj = 1.25597899e+04 Iter 40: Inner Its = 14 KKT violation = 4.438471e-03, nViolations = 0, obj = 1.25597899e+04 Iter 50: Inner Its = 12 KKT violation = 3.874232e-03, nViolations = 0, obj = 1.25597899e+04 Iter 60: Inner Its = 13 KKT violation = 3.496665e-03, nViolations = 0, obj = 1.25597899e+04 Iter 70: Inner Its = 12 KKT violation = 2.484731e-03, nViolations = 0, obj = 1.25597899e+04 Iter 80: Inner Its = 12 KKT violation = 1.799994e-03, nViolations = 0, obj = 1.25597899e+04 Iter 90: Inner Its = 13 KKT violation = 1.321612e-03, nViolations = 0, obj = 1.25597899e+04 Iter 100: Inner Its = 12 KKT violation = 9.791504e-04, nViolations = 0, obj = 1.25597899e+04 Iter 110: Inner Its = 12 KKT violation = 7.306154e-04, nViolations = 0, obj = 1.25597899e+04 Iter 120: Inner Its = 12 KKT violation = 5.474619e-04, nViolations = 0, obj = 1.25597899e+04 Iter 130: Inner Its = 12 KKT violation = 4.123053e-04, nViolations = 0, obj = 1.25597899e+04 Iter 140: Inner Its = 12 KKT violation = 3.103147e-04, nViolations = 0, obj = 1.25597899e+04 Iter 150: Inner Its = 12 KKT violation = 2.344776e-04, nViolations = 0, obj = 1.25597899e+04 Iter 160: Inner Its = 12 KKT violation = 1.776971e-04, nViolations = 0, obj = 1.25597899e+04 Iter 170: Inner Its = 12 KKT violation = 1.349647e-04, nViolations = 0, obj = 1.25597899e+04 Iter 180: Inner Its = 12 KKT violation = 1.024869e-04, nViolations = 0, obj = 1.25597899e+04 Exiting because all subproblems reached KKT tol. =========================================== Final f = 1.255979e+04 Final least squares fit = 5.232300e-01 Final KKT violation = 9.9944338e-05 Total inner iterations = 2719 Total execution time = 1.56 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'mu' (multiplicative updates with adjustments) stoptol = 0.0001, stoptime = 1e+06 maxiters = 200, maxinneriters = 10 epsDivZero = 1e-10, kappa = 50, kappatol = 1e-10 Iter 10: Inner Its = 30 KKT violation = 3.322665e-01, nViolations = 3, obj = 1.43503679e+04 Iter 20: Inner Its = 30 KKT violation = 6.261468e-01, nViolations = 3, obj = 1.25694191e+04 Iter 30: Inner Its = 30 KKT violation = 1.165293e-02, nViolations = 0, obj = 1.25592873e+04 Iter 40: Inner Its = 13 KKT violation = 1.242409e-03, nViolations = 0, obj = 1.25592868e+04 Iter 50: Inner Its = 12 KKT violation = 9.159682e-04, nViolations = 0, obj = 1.25592868e+04 Iter 60: Inner Its = 12 KKT violation = 6.805259e-04, nViolations = 0, obj = 1.25592868e+04 Iter 70: Inner Its = 12 KKT violation = 5.088195e-04, nViolations = 0, obj = 1.25592868e+04 Iter 80: Inner Its = 12 KKT violation = 3.806999e-04, nViolations = 0, obj = 1.25592868e+04 Iter 90: Inner Its = 12 KKT violation = 2.862512e-04, nViolations = 0, obj = 1.25592868e+04 Iter 100: Inner Its = 12 KKT violation = 2.160233e-04, nViolations = 0, obj = 1.25592868e+04 Iter 110: Inner Its = 12 KKT violation = 1.625292e-04, nViolations = 0, obj = 1.25592868e+04 Iter 120: Inner Its = 12 KKT violation = 1.224492e-04, nViolations = 0, obj = 1.25592868e+04 Exiting because all subproblems reached KKT tol. =========================================== Final f = 1.255929e+04 Final least squares fit = 5.232290e-01 Final KKT violation = 9.9963935e-05 Total inner iterations = 2203 Total execution time = 1.23 secs CP_APR: Tensor size = [100 80 60], Approximation Rank = 5 Tensor type: sparse with 10450 (2.2%) nonzeros alg = 'mu' (multiplicative updates with adjustments) stoptol = 0.0001, stoptime = 1e+06 maxiters = 200, maxinneriters = 10 epsDivZero = 1e-10, kappa = 50, kappatol = 1e-10 Iter 10: Inner Its = 30 KKT violation = 3.324546e-01, nViolations = 1, obj = 1.43721467e+04 Iter 20: Inner Its = 30 KKT violation = 1.254772e-01, nViolations = 1, obj = 1.43639954e+04 Iter 30: Inner Its = 30 KKT violation = 9.974392e-02, nViolations = 1, obj = 1.43589038e+04 Iter 40: Inner Its = 30 KKT violation = 1.314063e-01, nViolations = 1, obj = 1.43533891e+04 Iter 50: Inner Its = 30 KKT violation = 1.138546e-01, nViolations = 1, obj = 1.43513304e+04 Iter 60: Inner Its = 30 KKT violation = 3.344027e-02, nViolations = 0, obj = 1.43507349e+04 Iter 70: Inner Its = 30 KKT violation = 3.036969e-02, nViolations = 0, obj = 1.43503299e+04 Iter 80: Inner Its = 30 KKT violation = 1.267476e-01, nViolations = 2, obj = 1.43497493e+04 Iter 90: Inner Its = 30 KKT violation = 5.061820e-01, nViolations = 1, obj = 1.43457793e+04 Iter 100: Inner Its = 30 KKT violation = 1.366917e-01, nViolations = 3, obj = 1.43430420e+04 Iter 110: Inner Its = 30 KKT violation = 2.003749e-02, nViolations = 0, obj = 1.43407603e+04 Iter 120: Inner Its = 30 KKT violation = 1.892862e-02, nViolations = 0, obj = 1.43397194e+04 Iter 130: Inner Its = 30 KKT violation = 2.069699e-02, nViolations = 0, obj = 1.43394426e+04 Iter 140: Inner Its = 30 KKT violation = 1.989947e-02, nViolations = 0, obj = 1.43393698e+04 Iter 150: Inner Its = 30 KKT violation = 1.591972e-02, nViolations = 0, obj = 1.43393514e+04 Iter 160: Inner Its = 25 KKT violation = 1.597734e-02, nViolations = 0, obj = 1.43393468e+04 Iter 170: Inner Its = 23 KKT violation = 1.011655e-02, nViolations = 0, obj = 1.43393454e+04 Iter 180: Inner Its = 22 KKT violation = 6.328913e-03, nViolations = 0, obj = 1.43393450e+04 Iter 190: Inner Its = 21 KKT violation = 4.910786e-03, nViolations = 0, obj = 1.43393448e+04 Iter 200: Inner Its = 13 KKT violation = 4.885278e-03, nViolations = 0, obj = 1.43393447e+04 =========================================== Final f = 1.433934e+04 Final least squares fit = 5.154573e-01 Final KKT violation = 4.8852779e-03 Total inner iterations = 5629 Total execution time = 3.58 secs
Comparing results
We can see that all methods can find a reasonable solution, though convergence speed and final accuracy might differ. The 'pqnr' and 'pdnr' methods are generally more sophisticated and may converge to a better solution or faster in terms of outer iterations, while 'mu' iterations are typically cheaper.
For this particular problem and random initialization:
fprintf('Factor Match Score (PQNR): %.4f\n', factor_match_score_pqnr); fprintf('Factor Match Score (PDNR): %.4f\n', factor_match_score_pdnr); fprintf('Factor Match Score (MU): %.4f\n', factor_match_score_mu);
Factor Match Score (PQNR): 0.9744 Factor Match Score (PDNR): 0.9744 Factor Match Score (MU): 0.9744
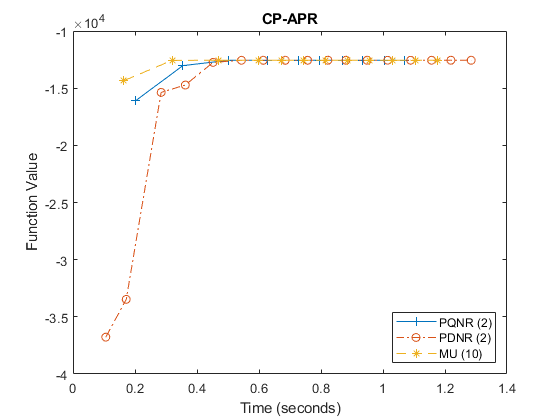
Visualize the results
The function values are only computed when printing the output to the screen. The legend indicates the method and, in parentheses, how frequently the objective value was recorded.
figure(1); clf; tf = ~isnan(output_pqnr.fnVals); plot(output_pqnr.times(tf), 1-output_pqnr.fnVals(tf),'-+','DisplayName','PQNR (2)'); hold on; tf = ~isnan(output_pdnr.fnVals); plot(output_pdnr.times(tf), 1-output_pdnr.fnVals(tf),'-.o', 'DisplayName','PDNR (2)'); tf = ~isnan(output_mu.fnVals); plot(output_mu.times(tf), 1-output_mu.fnVals(tf),'--*', 'DisplayName','MU (10)'); hold off; title('CP-APR'); xlabel('Time (seconds)'); ylabel('Function Value'); legend('Location','SouthEast')