Alternating randomized least squares for CP Decomposition
The function cp_arls computes an estimate of the best rank-R CP model of a tensor X using alternating randomized least-squares algorithm. The input X must be a (dense) tensor. The output CP model is a ktensor.
The method is a randomized version of the alternating least squares (ALS) method, which is a popular method for computing CP decompositions. The randomized method is generally faster and more robust than the standard ALS method for very large tensors. Each iteration, it samples a subset of the rows in the overdetermined least squares problem and solves that smaller problem.
The fit value is calculated as 1 minus the relative error, i.e., 1 - norm(X-full(M))/norm(X) where X is the tensor and M is the model. This is generally interpreted as the quality of the fit, with values closer to 1 indicating a better fit. Because computing the fit is expensive, this method uses a sampling strategy to estimate the fit value without computing it exactly.
The CP-ARLS method is described in the following reference:
- C. Battaglino, G. Ballard, T. G. Kolda. A Practical Randomized CP Tensor Decomposition. SIAM Journal on Matrix Analysis and Applications 39(2):876-901, 2018. https://doi.org/10.1137/17M1112303
Contents
- Set up a sample problem
- Running the CP-ARLS method
- Plot the fit trace
- Speed things up by skipping the initial mixing
- Comparing with CP-ALS
- How well does the approximate fit do?
- Varying epoch size
- Set up another sample problem
- Terminating once a desired fit is achieved
- Changing the number of function evaluation samples
- Change the number of sampled rows in least squares solve
Set up a sample problem
We set up an especially difficult and somewhat large sample problem that has high collinearity (0.9) and 1% noise. This is an example where the randomized method will generally outperform the standard method.
rng('default') %<- Setting random seed for reproducibility of this script sz = [200 300 400]; R = 5; ns = 0.01; coll = 0.9; info = create_problem('Size', sz, 'Num_Factors', R, 'Noise', ns, ... 'Factor_Generator', @(m,n) matrandcong(m,n,coll), ... 'Lambda_Generator', @ones); % Extract data and solution X = info.Data; M_true = info.Soln;
Running the CP-ARLS method
Running the method is essentially the same as using CP-ALS, feed the data matrix and the desired rank. Note that the iteration is of the form NxN which is the number of epochs x the number of iterations per epoch. The default number of iterations per epoch is 50. At the end of each epoch, we check the convergence criteria. Because this is a randomized method, we do not achieve strict decrease in the objective function. Instead, we look at the number of epochs without improvement (newi) and exit when this crosses the predefined tolerance (newitol), which defaults to 5. It is important to note that the fit values that are reported are approximate, so this is why it is denoted by f~ rather than just f.
tic [M1, ~, out1] = cp_arls(X,R); time1 = toc; scr1 = score(M1,M_true); fprintf('\n*** Results for CP-ARLS (with mixing) ***\n'); fprintf('Time (secs): %.3f\n', time1) fprintf('Score (max=1): %.3f\n', scr1);
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 5 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 16384 Iter 10x5: f~ = 9.885167e-01 newi = 0 Iter 20x5: f~ = 9.892368e-01 newi = 0 Iter 30x5: f~ = 9.894751e-01 newi = 0 Iter 40x5: f~ = 9.896253e-01 newi = 4 Iter 50x5: f~ = 9.897155e-01 newi = 1 Iter 60x5: f~ = 9.897859e-01 newi = 0 Iter 70x5: f~ = 9.897710e-01 newi = 2 Iter 73x5: f~ = 9.898088e-01 newi = 5 *** Results for CP-ARLS (with mixing) *** Time (secs): 2.850 Score (max=1): 0.956
Plot the fit trace
If the trace is enabled, we can plot the fit trace to see how the fit changes over time. This is useful to see how the method converges and to identify any potential issues with the optimization process.
tic [M, ~, out] = cp_arls(X,R,'trace',true); runtime = toc; scr = score(M,M_true); fprintf('\n*** Results for CP-ARLS (with trace) ***\n'); fprintf('Time (secs): %.3f\n', runtime) fprintf('Score (max=1): %.3f\n', scr); figure(1);clf; plot(out.time_trace+out.init_time, out.fit_trace, '-o'); xlabel('Time (seconds)'); ylabel('Fit (1 - relative error)'); title(sprintf('CP-ARLS Fit Trace (R=%d, Time=%.2f seconds)', R, runtime));
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 5 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 16384 Iter 10x5: f~ = 9.887588e-01 newi = 0 Iter 20x5: f~ = 9.892722e-01 newi = 0 Iter 30x5: f~ = 9.894888e-01 newi = 0 Iter 40x5: f~ = 9.896591e-01 newi = 0 Iter 50x5: f~ = 9.897628e-01 newi = 4 Iter 51x5: f~ = 9.897571e-01 newi = 5 *** Results for CP-ARLS (with trace) *** Time (secs): 1.953 Score (max=1): 0.903

Speed things up by skipping the initial mixing
The default behavior is to mix the data in each mode using an FFT and diagonal random +/-1 matrix. This may add substantial preprocessing time, though it helps to ensure that the method converges. Oftentimes, such as with randomly-generated data, the mixing is not necessary.
tic [M2, ~, out2] = cp_arls(X,R,'mix',false); time2 = toc; scr2 = score(M2,M_true); fprintf('\n*** Results for CP-ARLS (no mix) ***\n'); fprintf('Time (secs): %.3f\n', time2) fprintf('Score (max=1): %.3f\n', scr2);
CP_ARLS (without mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 5 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 16384 Iter 10x5: f~ = 9.716017e-01 newi = 1 Iter 20x5: f~ = 9.886206e-01 newi = 0 Iter 30x5: f~ = 9.887216e-01 newi = 5 *** Results for CP-ARLS (no mix) *** Time (secs): 0.780 Score (max=1): 0.804
Comparing with CP-ALS
CP-ALS may be somewhat faster, especially since this is a relatively small problem, but it usually will not achieve as good of an answer in terms of the score.
tic; [M3, ~, out3] = cp_als(X,R,'maxiters',500,'printitn',10); time3 = toc; scr3 = score(M3,M_true); fprintf('\n*** Results for CP-ALS ***\n'); fprintf('Time (secs): %.3f\n', time3) fprintf('Score (max=1): %.3f\n', scr3);
CP_ALS (CP Alternating Least Squares): Tensor size: [200 300 400] Tensor type: tensor R = 5, maxiters = 500, tol = 1.000000e-04 dimorder = [1 2 3] init = random Iter 10: f = 9.566043e-01 f-delta = 4.0e-04 Iter 20: f = 9.583031e-01 f-delta = 1.1e-04 Iter 22: f = 9.585007e-01 f-delta = 9.6e-05 Final f = 9.585007e-01 *** Results for CP-ALS *** Time (secs): 0.492 Score (max=1): 0.313
How well does the approximate fit do?
It is possible to check the accuracy of the fit computation by having the code compute the true fit and the final solution, enabled by the truefit option.
[M4,~,out4] = cp_arls(X,R,'truefit',true);
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 5 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 16384 Iter 10x5: f~ = 9.738242e-01 newi = 0 Iter 20x5: f~ = 9.894454e-01 newi = 0 Iter 30x5: f~ = 9.894567e-01 newi = 3 Iter 32x5: f~ = 9.894580e-01 newi = 5 Final fit = 9.895512e-01 Final estimated fit = 9.895421e-01
Varying epoch size
It is possible to vary that number of iterations per epoch. Fewer iterations means that more time is spent checking for convergence and it may also be harder to detect as an single iteration can have some fluctuation and we are actually looking for the overall trend. In contrast, too many iterations means that the method won't realize when it has converged and may spend too much time computing.
tic M = cp_arls(X,R,'epoch',1,'newitol',20); toc fprintf('Score: %.4f\n',score(M,M_true));
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 1, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 20 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 16384 Iter 10x1: f~ = 9.708390e-01 newi = 0 Iter 20x1: f~ = 9.837763e-01 newi = 0 Iter 30x1: f~ = 9.862112e-01 newi = 0 Iter 40x1: f~ = 9.878606e-01 newi = 0 Iter 50x1: f~ = 9.884283e-01 newi = 0 Iter 60x1: f~ = 9.887198e-01 newi = 0 Iter 70x1: f~ = 9.888976e-01 newi = 0 Iter 80x1: f~ = 9.890788e-01 newi = 0 Iter 90x1: f~ = 9.891355e-01 newi = 0 Iter 100x1: f~ = 9.891999e-01 newi = 3 Iter 110x1: f~ = 9.893096e-01 newi = 0 Iter 120x1: f~ = 9.893742e-01 newi = 0 Iter 130x1: f~ = 9.894091e-01 newi = 0 Iter 140x1: f~ = 9.893893e-01 newi = 1 Iter 150x1: f~ = 9.894508e-01 newi = 5 Iter 160x1: f~ = 9.894768e-01 newi = 9 Iter 170x1: f~ = 9.895068e-01 newi = 3 Iter 180x1: f~ = 9.895040e-01 newi = 7 Iter 190x1: f~ = 9.895315e-01 newi = 5 Iter 200x1: f~ = 9.895652e-01 newi = 6 Iter 210x1: f~ = 9.895402e-01 newi = 4 Iter 220x1: f~ = 9.896326e-01 newi = 4 Iter 230x1: f~ = 9.896330e-01 newi = 3 Iter 240x1: f~ = 9.895865e-01 newi = 13 Iter 247x1: f~ = 9.896514e-01 newi = 20 Elapsed time is 2.114009 seconds. Score: 0.9180
tic M = cp_arls(X,R,'epoch',200,'newitol',3,'printitn',2); toc fprintf('Score: %.4f\n',score(M,M_true));
CP-ARLS (with mixing): Print every 2 epochs, Epoch size: 200, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 3 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 16384 Iter 2x200: f~ = 9.896270e-01 newi = 0 Iter 4x200: f~ = 9.896865e-01 newi = 0 Iter 6x200: f~ = 9.896158e-01 newi = 2 Iter 7x200: f~ = 9.896663e-01 newi = 3 Elapsed time is 10.234171 seconds. Score: 0.9868
Set up another sample problem
We set up another problem with 10% noise, but no collinearity.
sz = [200 300 400]; R = 5; ns = 0.10; info = create_problem('Size', sz, 'Num_Factors', R, 'Noise', ns, ... 'Factor_Generator', @rand,'Lambda_Generator', @ones); % Extract data and solution X = info.Data; M_true = info.Soln;
Terminating once a desired fit is achieved
If we know the noise level is 10%, we would expect a fit of 0.90 at best. So, we can set a threshold that is close to that and terminate as soon as we achieve that accuracy. Since detecting convergence is hard for a randomized method, this can lead to speed ups. However, if the fit is not high enough, the accuracy may suffer consequently.
M = cp_arls(X,R,'newitol',20,'fitthresh',0.895,'truefit',true); fprintf('Score: %.4f\n',score(M,M_true));
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 20 Stop if f (approx) greater than: 0.895 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 16384 Iter 6x5: f~ = 8.968036e-01 newi = 0 Final fit = 8.968118e-01 Final estimated fit = 8.968036e-01 Score: 0.9523
Changing the number of function evaluation samples
The function evaluation is approximate and based on sampling the number of entries specified by nsampfit. If this is too small, the samples will not be accurate enough. If this is too large, the computation will take too long. The default is 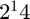 , which should generally be sufficient. It may sometimes be possible to use smaller values. The same sampled entries are used for every convergence check --- we do not resample to check other entries.
, which should generally be sufficient. It may sometimes be possible to use smaller values. The same sampled entries are used for every convergence check --- we do not resample to check other entries.
M = cp_arls(X,R,'truefit',true,'nsampfit',100); fprintf('Score: %.4f\n',score(M,M_true));
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 5 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 117 Entry samples for approximate fit: 100 Iter 10x5: f~ = 8.915405e-01 newi = 3 Iter 12x5: f~ = 8.935686e-01 newi = 5 Final fit = 8.948320e-01 Final estimated fit = 8.958579e-01 Score: 0.9565
Change the number of sampled rows in least squares solve
The default number of sampled rows for the least squares solves is ceil(10*R*log2( R )). This seemed to work well in most tests, but this can be varied higher or lower. For R=5, this means we sample 117 rows per solve. The rows are different for every least squares problem. Let's see what happens if we reduce this to 10.
M = cp_arls(X,R,'truefit',true,'nsamplsq',10); fprintf('Score: %.4f\n',score(M,M_true));
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 5 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 10 Entry samples for approximate fit: 16384 Iter 10x5: f~ = 6.832712e-01 newi = 5 Final fit = 7.584470e-01 Final estimated fit = 7.598184e-01 Score: 0.3294
What if we use 25?
M = cp_arls(X,R,'truefit',true,'nsamplsq',25); fprintf('Score: %.4f\n',score(M,M_true));
CP-ARLS (with mixing): Print every 10 epochs, Epoch size: 5, Max epochs: 1000 Improvement tolerance: 0.000e+00 Stop after this many iterations without improvement: 5 Stop if f (approx) greater than: 1.000 Initial guess: char Least-squares row samples: 25 Entry samples for approximate fit: 16384 Iter 10x5: f~ = 8.745437e-01 newi = 3 Iter 16x5: f~ = 8.787478e-01 newi = 5 Final fit = 8.818711e-01 Final estimated fit = 8.817138e-01 Score: 0.8576
